Viral Vector Titer Methods: Capsid, Genome & Infectious Titers Explained
When someone asks, “How much vector do we have?” the answer is never as simple as a single number.
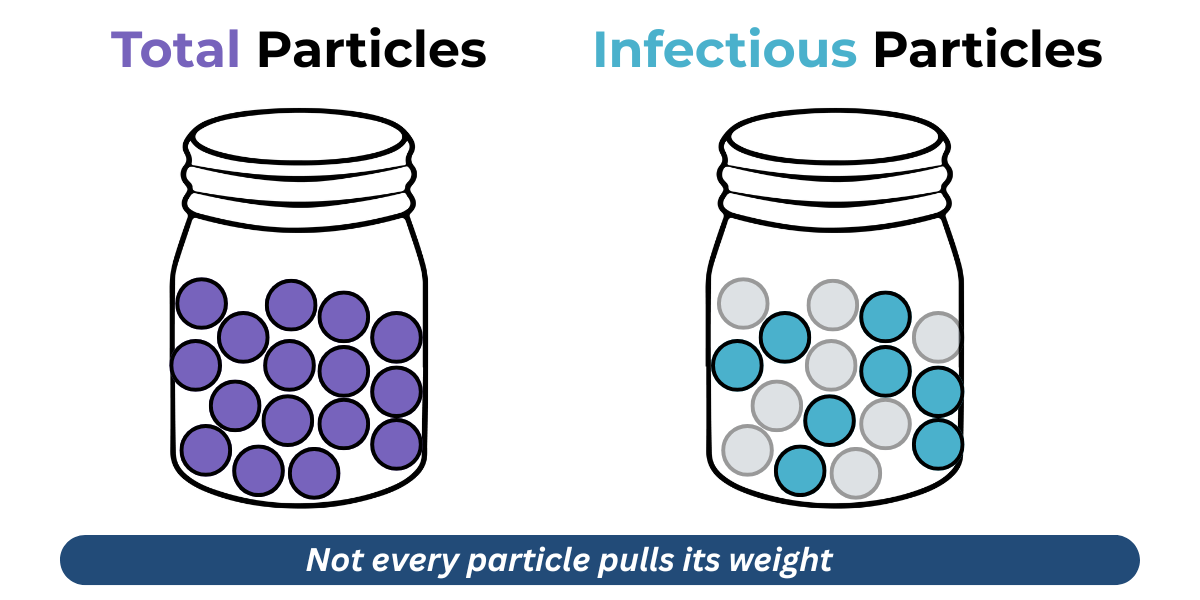
That’s because not all viral particles are created equal. Some are functional, delivering their genetic payload like champions. Others are just along for the ride - empty shells or defective copies that look like the part but don’t do the job.
Understanding this distinction is critical. A batch may look abundant, but what really matters is how many particles are truly infectious, and how that relates to the total number of viral particles (Particle:Infectivity ratio).
Includes a one-page overview of capsid, genome, and infectious titer assays.
3 Ways We Measure Yield
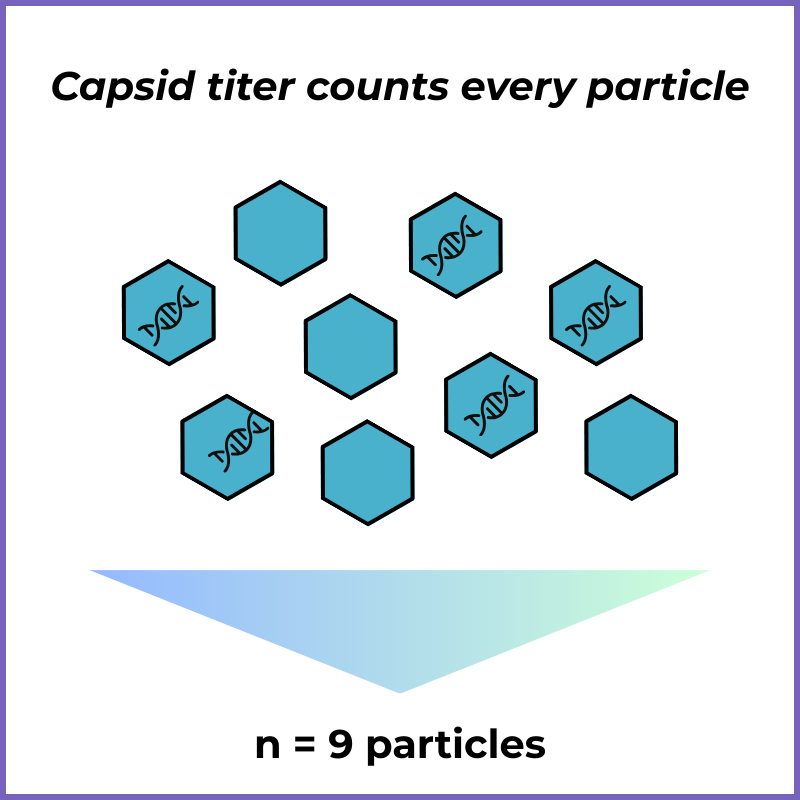
Capsid Titer (Protein-based)
What it is: Measures how many viral “shells” (capsids) are present, regardless of what’s inside.
Example: For lentiviral vectors, a common method is the p24 ELISA, which detects a structural protein in the viral capsid.
What it tells you: “How many virus particles were made?”
Limitations: Doesn’t distinguish between full vs. empty particles, or between infectious vs. defective ones.
Genome Titer
What it is: Counts how many viral genomes are packaged.
Example: For AAV, qPCR against ITR sequence is standard.
What it tells you: “How many genomes are present inside capsids?” Since each viral particle packages a specific number of genome copies (1 for AAV, 2 for Lentiviral vector), you can use this to determine how many viral particles you have.
Limitations: Can’t tell if the packaged genome is intact, or if the virus is functional once it enters a cell.
(See Kelley-Clarke et al. 2014 for a detailed description of a lentiviral vector genome titer method.)
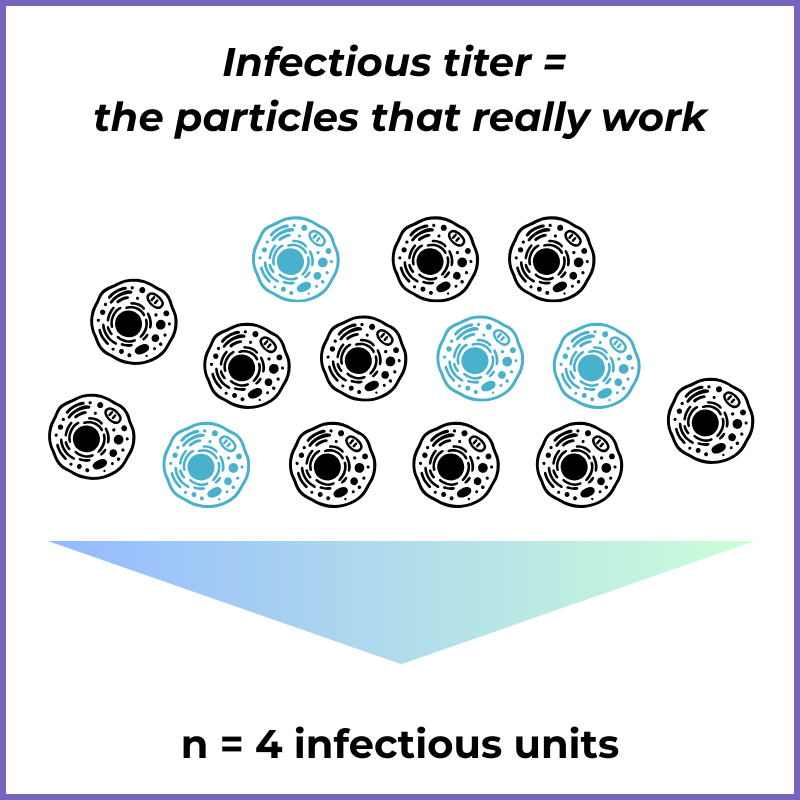
Infectious Titer (Function-based)
What it is: Measures the fraction of particles that actually infect cells and deliver the genetic payload.
Example: For lentiviral vectors, cell-based assays with a qPCR or flow-based readout can directly assess functionality.
What it tells you: “How many particles are doing the job we care about?”
Limitations: Cell-based assays are more variable and have longer turnaround time than ELISA or PCR-based methods. In addition, the assay design must be specific to the vector system and payload.
(See Kelley-Clarke et al., 2016 for a detailed lentiviral vector infectious titer assay method.)
Why They Don’t Match
Even if you measure the same batch of vector by all three methods, the numbers will differ - sometimes dramatically. Why?
Not every capsid contains a genome
Not every genome is full intact or packaged correctly
Not every correctly packaged viral particle can infect/transduce a cell
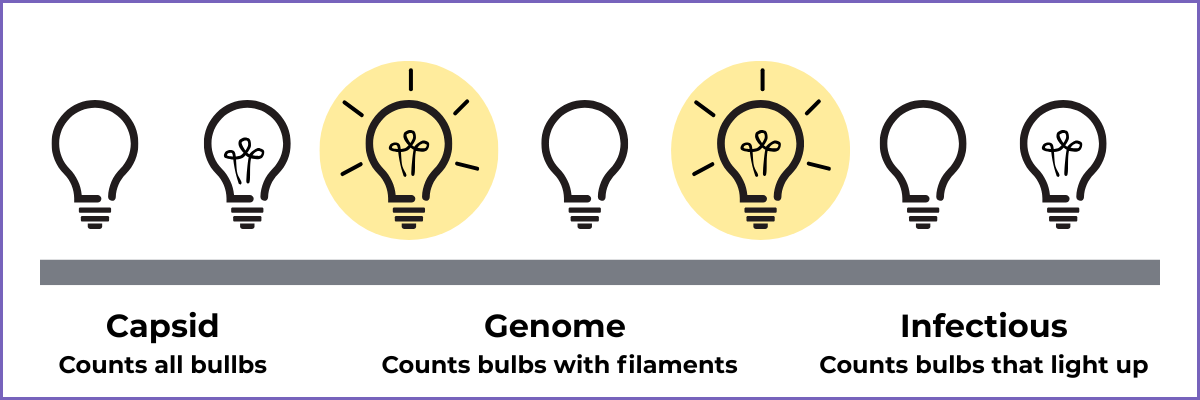
Think of it like a factory producing old-school light bulbs:
Capsid titer = How many bulbs roll off the line
Genome titer = How many bulbs have filaments inside
Infectious titer = How many bulbs can actually light up
Why It Matters
The difference between particle counts and infectious units isn’t just technical hair-splitting. It has real-world consequences:
Dosing: Patients need infectious particles, not just total particles. A misleading count could mean under- or over-dosing.
Safety: Non-infectious particles can still trigger immune responses, complicating therapy.
Manufacturing Strategy: High particle-to-infectivity ratios can point to manufacturing inefficiencies that should be addressed upstreams (e.g. by varying plasmid ratios or transfection conditions).
Key Takeaways
Not all viral particles are infectious
Different assays tell different parts of the story
Robust viral vector analytics are essential for accurate dosing, safety, and regulatory success
Getting in Touch
Have a question or want to explore working together?